Vehicular Emissions INventories (VEIN)

TODO
- Include speed functions with Fortran
- Include CB6
- Add EF from HBEFA?
- See issues GitHub
- Second edition of my book
System requirements
vein imports functions from spatial packages listed below. In order to install these packages, firstly the user must install the requirements mentioned here.
Installation
GitHub
remotes::install_github("atmoschem/vein")or if you have a 32 bits machine
install_github("atmoschem/vein",
INSTALL_opts = "--no-multiarch")Run with a project
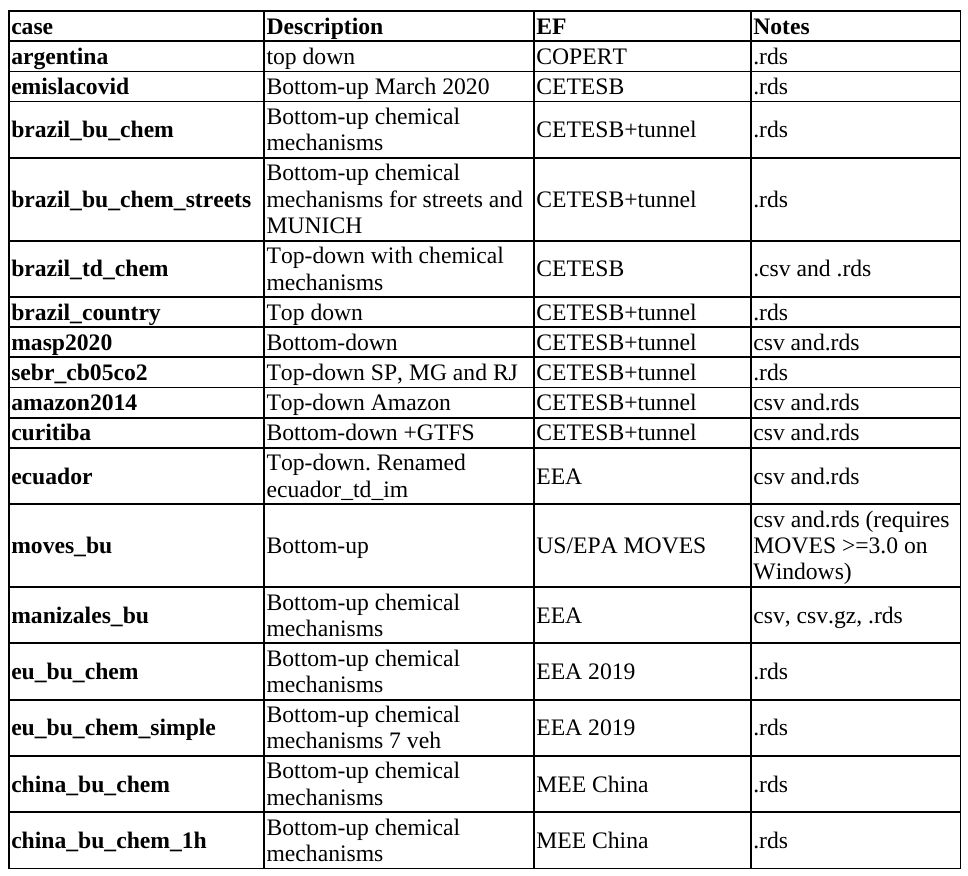
Use the function get_project and read the documentation, there you can see more projects as well.
library(vein)
?get_project
get_project(directory = "awesome_city")The structure of the new directory “awesome_city” is:
awesome_city
├── config
│ ├── clean.R
│ ├── config.R
│ ├── inventory.xlsx
│ └── packages.R
├── main.R
├── main.Rproj
├── network
│ ├── net.gpkg
│ └── net.rds
├── scripts
│ ├── evaporatives.R
│ ├── exhaust.R
│ ├── fuel_eval.R
│ ├── net.R
│ ├── pavedroads.R
│ ├── plots.R
│ ├── post.R
│ ├── traffic.R
│ └── wrf.R
└── wrf
└── wrfinput_d02You have to open the file main.Rproj with Rstudio and then open and run main.R
To run main.R you will need these extra packages:
- ggplot2
- readxl
- eixport (If you plan to generate WRF Chem emissions file)
If you do not have them already, you can install:
install.packages(c("ggplot2", "readxl", "eixport"))Check the projects here
Citation
If you use VEIN, please, cite it (BIBTEX, ENDNOTE):
Ibarra-Espinosa, S., Ynoue, R., O’Sullivan, S., Pebesma, E., Andrade, M. D. F., and Osses, M.: VEIN v0.2.2: an R package for bottom-up vehicular emissions inventories, Geosci. Model Dev., 11, 2209-2229, https://doi.org/10.5194/gmd-11-2209-2018, 2018.
@article{gmd-11-2209-2018,
author = {Ibarra-Espinosa, S. and Ynoue, R. and O'Sullivan, S. and Pebesma, E. and Andrade, M. D. F. and Osses, M.},
title = {VEIN v0.2.2: an R package for bottom--up vehicular emissions inventories},
journal = {Geoscientific Model Development},
volume = {11},
year = {2018},
number = {6},
pages = {2209--2229},
url = {https://gmd.copernicus.org/articles/11/2209/2018/},
doi = {10.5194/gmd-11-2209-2018}
}Communications, doubts etc
- Earth-Sciences on Stackoverflow, tag vein-r-package
- Drop me an email sergio.ibarraespinosa@colorado.edu or zergioibarra@hotmail.com (你好中国朋友 - Hello Chinese friends!)
- Check the group on GoogleGroups Group.
Issues
If you encounter any issues while using VEIN, please submit your issues to: https://github.com/atmoschem/vein/issues/ If you have any suggestions just let me know to sergio.ibarra@usp.br.
Contributing
Please, read this guide. Contributions of all sorts are welcome, issues and pull requests are the preferred ways of sharing them. When contributing pull requests, please follow the Google’s R Style Guide.
Note for non-english and anaconda users
Sometimes you need to install R and all dependencies and a way for doing that is using anaconda. Well, as my system is in portuguese, after installing R using anaconda it changed the decimal character to ‘,’. In order to change it back to english meaning decimal separator as ‘.’, I added this variable into the .bashrc
More details on StackOverflow